Chapter 13 Multiple Regression
In this chapter, we consider the case where we have a single numeric response variable and multiple explanatory variables. There is a natural tension between explaining the response variable well and keeping the model simple. Our main purpose will be to understand this tension and find a balance between the two competing goals.
13.1 Two explanatory variables
We consider housing sales data from King County, WA, which includes the city of Seattle.
The data set houses in the fosdata package contains a record of
every house sold in King County from May, 2014 through May, 2015. For each house, the data includes the sale price along
with many variables describing the house and its location.
Our goal is to model the sale price on the other variables.
There are many variables that we could use to do this, but in this section we will only consider two of them; namely, the square footage in the house (sqft_living) and the size of the lot (sqft_lot). To keep the size of the problem small, we restrict to the urban ZIP code 98115.
houses_98115 <- fosdata::houses %>%
filter(zipcode == "98115") %>%
select(price, sqft_living, sqft_lot)There are now 583 observations. We start by doing some summary statistics and visualizations.
## price sqft_living sqft_lot
## Min. : 200000 Min. : 620 Min. : 864
## 1st Qu.: 456750 1st Qu.:1270 1st Qu.: 4080
## Median : 567000 Median :1710 Median : 5300
## Mean : 619900 Mean :1835 Mean : 5444
## 3rd Qu.: 719000 3rd Qu.:2285 3rd Qu.: 6380
## Max. :2300000 Max. :5770 Max. :30122It appears that the variables may be right-skew. Let’s compute the skewness:
## price sqft_living sqft_lot
## 2.0482534 0.9114968 2.9309820The variables price and sqft_lot are right-skew and the variable sqft_living is moderately right-skew. Let’s look at histograms and scatterplots.
library(tidyr) # for pivot_longer
houses_98115 %>%
pivot_longer(cols = c(price, sqft_living, sqft_lot)) %>%
ggplot(aes(value)) +
geom_histogram(bins = 15) +
facet_wrap(~name, scales = "free_x")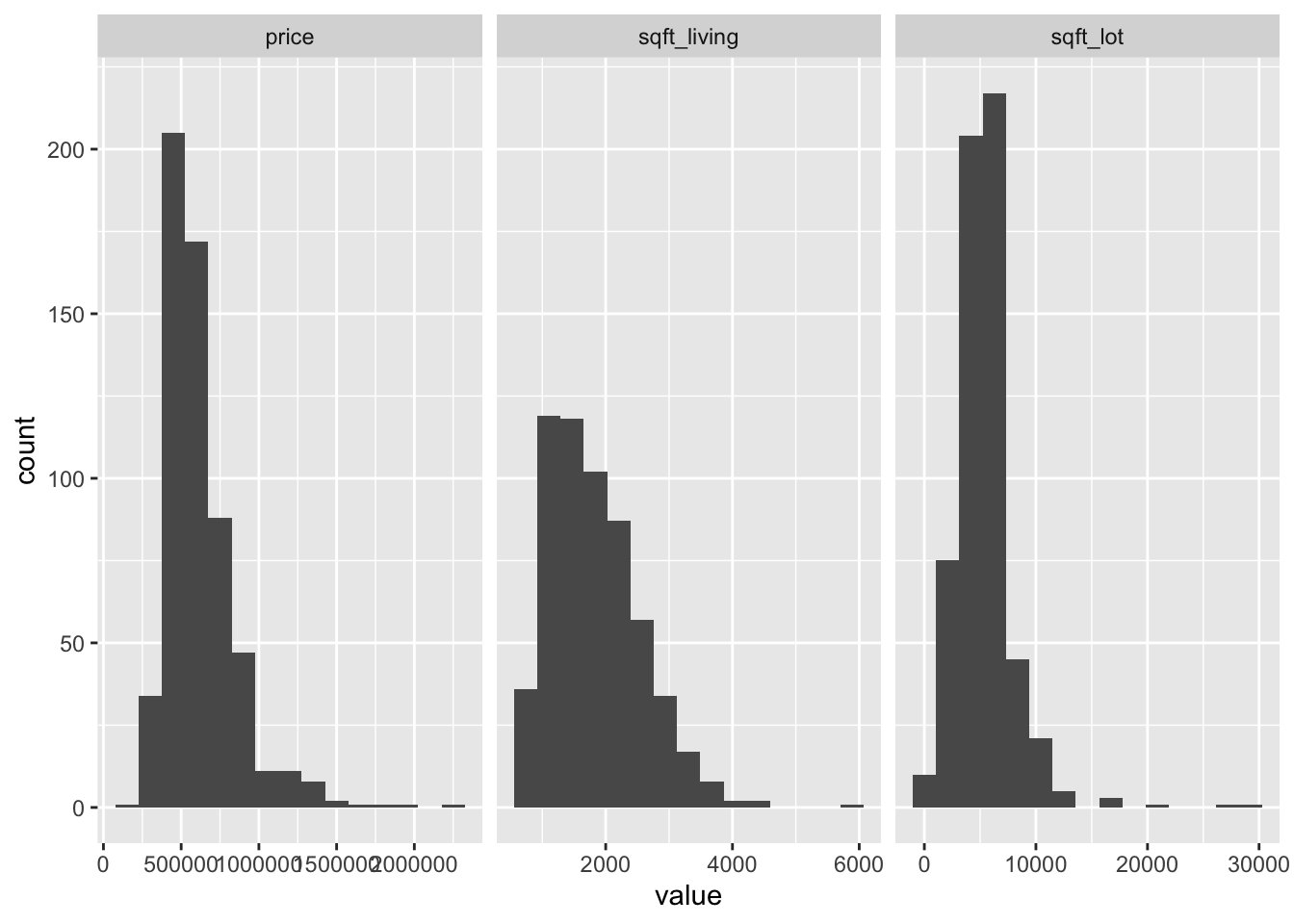
Now let’s look at two scatterplots.
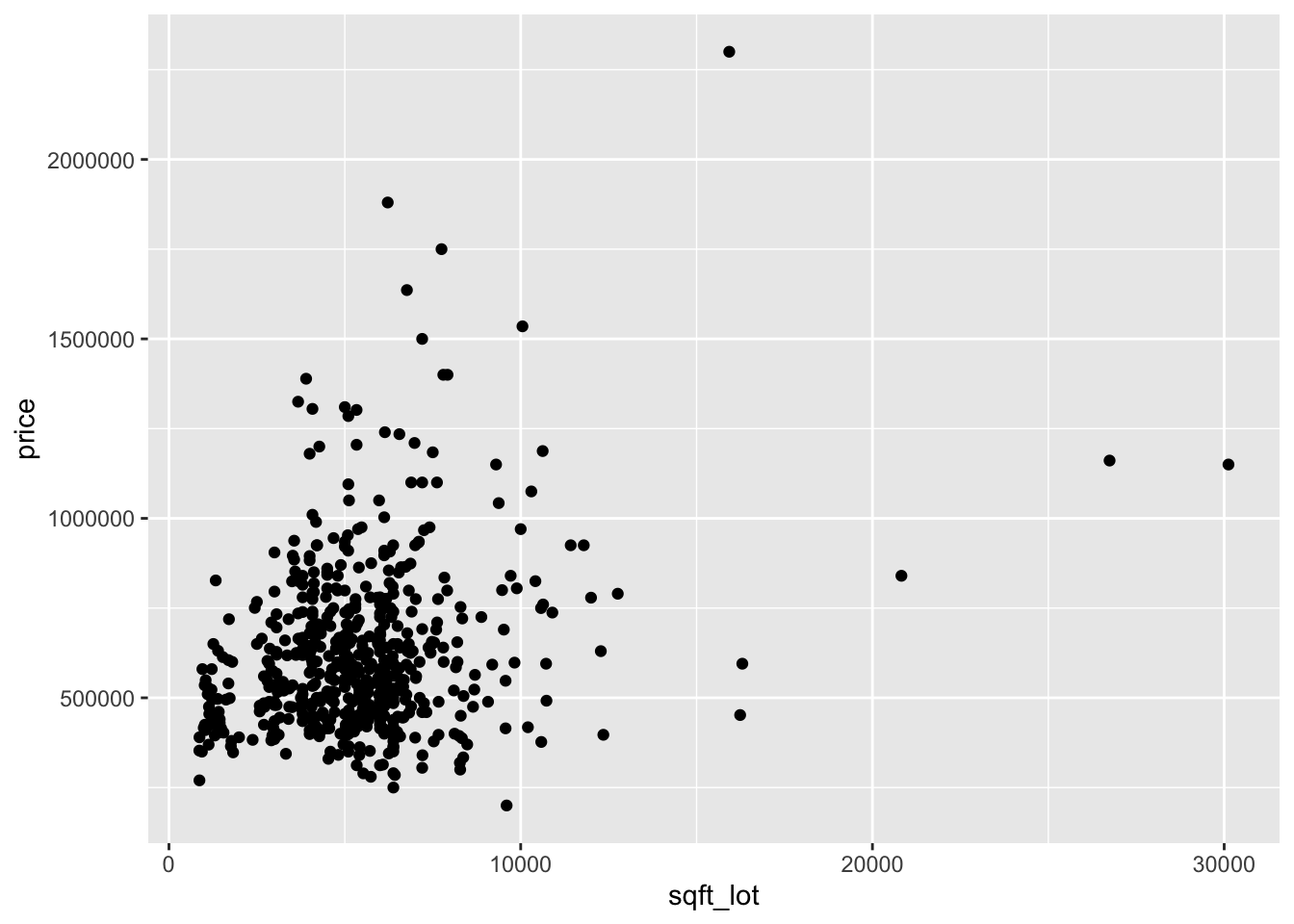
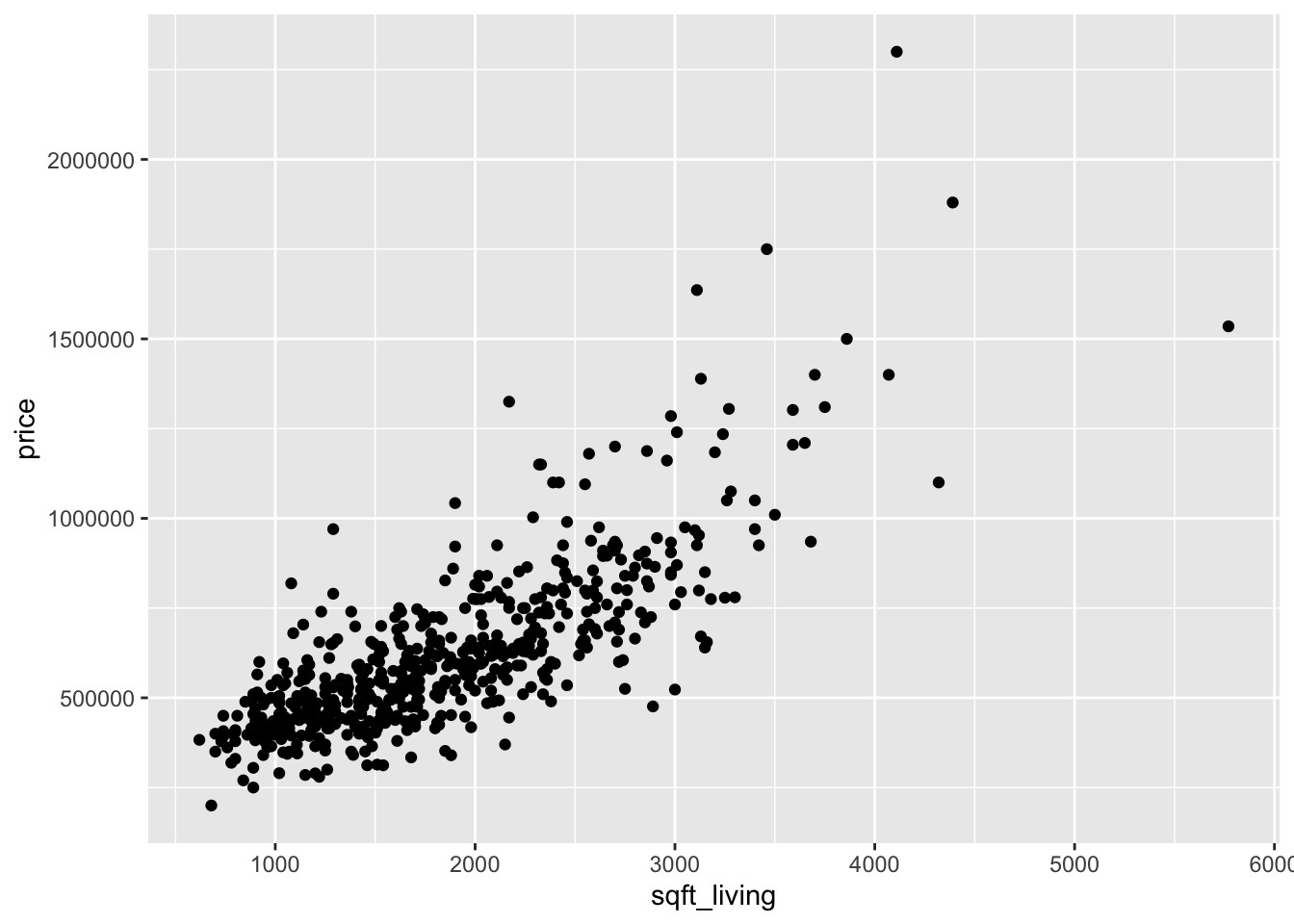
Looking at the two plots separately, it appears that the variance of price is not constant across the explanatory variables. Combining this with the fact that the variables are skew, we take logs of all three variables:
log_houses <- fosdata::houses %>%
filter(zipcode == "98115") %>%
mutate(
log_price = log(price),
log_sqft_living = log(sqft_living),
log_sqft_lot = log(sqft_lot)
) %>%
select(starts_with("log"))The scatterplots are improved, though log of price versus log of lot size does not look great.
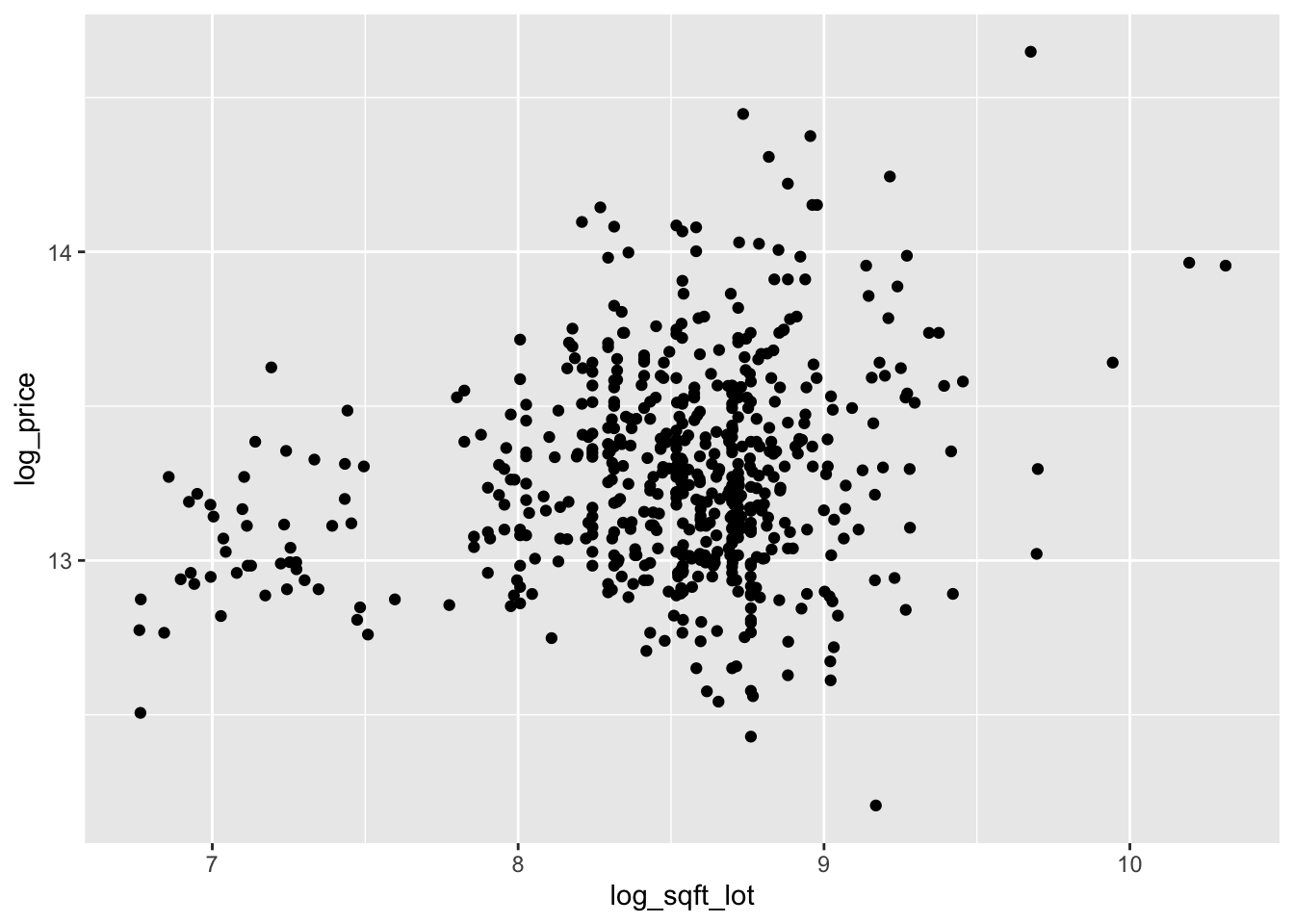
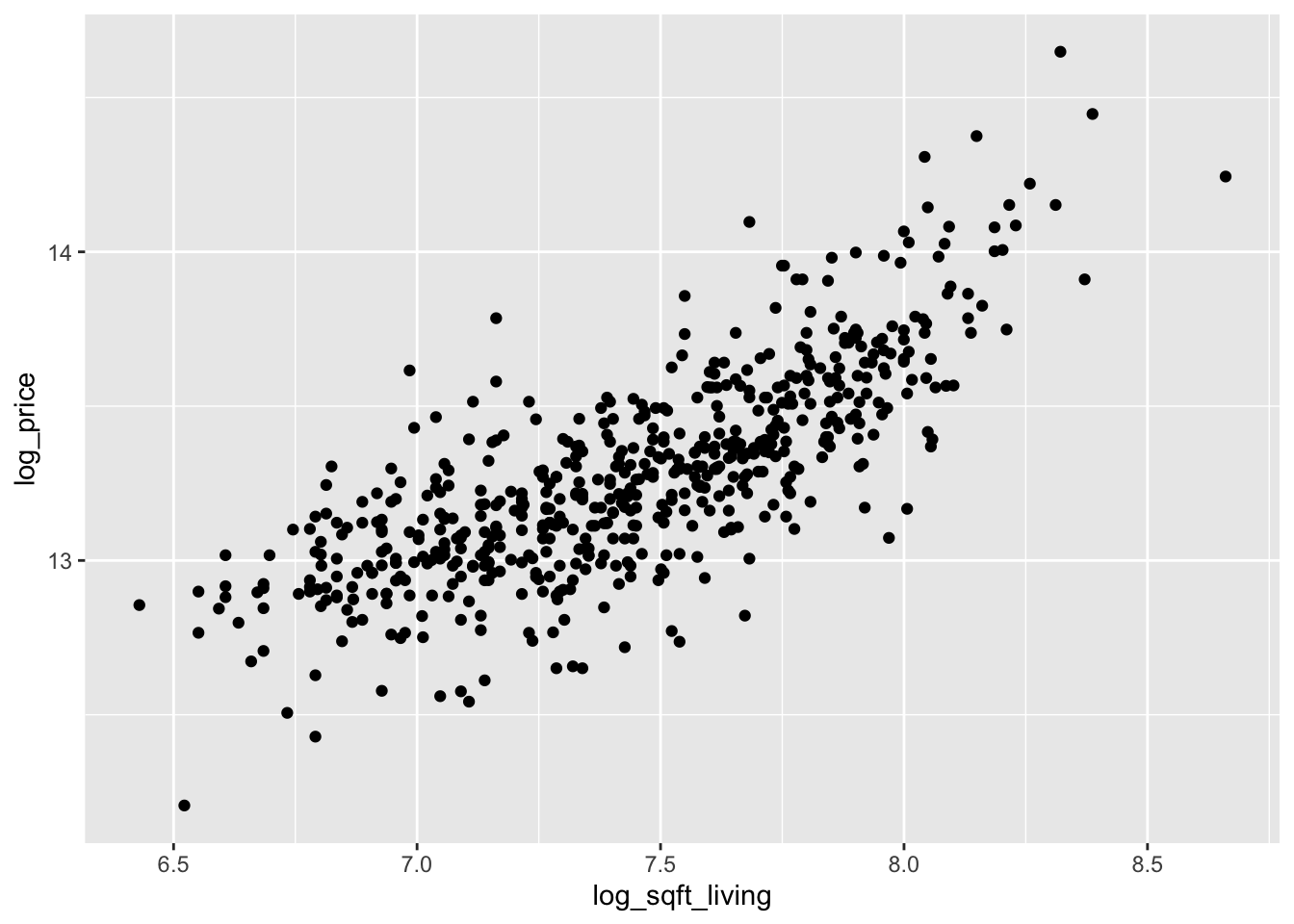
We continue by modeling the log of price on the log of the two explanatory variables.
With only two explanatory variables, it is still possible to visualize the relationship.
For scatterplots in three dimensions, we recommend plotly.
We will not be utilizing all of the features of plotly, so we do not go into detail. Plotly plots are interactive by default, so we recommend typing running the code below in R.
plotly::plot_ly(log_houses,
x = ~log_sqft_living,
y = ~log_sqft_lot,
z = ~log_price,
type = "scatter3d",
marker = list(size = 2)
)Looking at the scatterplot, it seems reasonable that a plane could model the relationship between \(x\), \(y\) and \(z\). How would we find the equation of the plane that is the best fit? Well, a plane has an equation of the form \(ax + by + cz = d\), and we are assuming that there is some dependence on \(z\), so \(c\not=0\). Therefore, we can write the equation of the plane as \(z = \beta_0 + \beta_1 x + \beta_2 y\) for some choices of \(\beta_0, \beta_1\) and \(\beta_2\). (In terms of the original equation, \(\beta_0 = d/c,\, \beta_1 = -b/c,\, \beta_2 = -a/c\).) For each \(\beta_0, \beta_1\) and \(\beta_2\) we can predict the values of \(z\) at the values of the explanatory variables by computing \(\hat z_i = \beta_0 + \beta_1 x_i + \beta_2 y_i\). The associated error is \(\hat z_i - z_i\), and we find the values of \(\beta_0, \beta_1\) and \(\beta_2\) that minimize the sum of squares of the error.
Model: Our model is \[ z_i = \beta_0 + \beta_1 x_i + \beta_2 y_i + \epsilon_i \] where \(\beta_0, \beta_1\) and \(\beta_2\) are unknown constants, and \(\epsilon_i\) are independent, identically distributed normal random variables. The \(i\)th response is given by \(z_i\), and the \(i\)th predictors are \(x_i\) and \(y_i\).
For example, if \(\beta_0 = \beta_1 = \beta_2 = 1\), then we can compute the sum of squared error via
beta_0 <- 1
beta_1 <- 1
beta_2 <- 1
log_houses %>%
mutate(
z_hat = beta_0 + beta_1 * log_sqft_living + beta_2 * log_sqft_lot,
resid = log_price - z_hat
) %>%
summarize(SSE = sum(resid^2))## SSE
## 1 7961.38By hand, adjust the values of \(\beta_0, \beta_1\) and \(\beta_2\) in the code above to try and minimize the SSE
We can find the values of \(\beta_0, \beta_1\) and \(\beta_2\) that minimize the SSE using lm. The format is very similar to the single explanatory variable case.
##
## Call:
## lm(formula = log_price ~ log_sqft_living + log_sqft_lot, data = log_houses)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.62790 -0.12085 -0.00189 0.12904 0.75567
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 8.14199 0.19922 40.87 <2e-16 ***
## log_sqft_living 0.65130 0.02303 28.29 <2e-16 ***
## log_sqft_lot 0.03422 0.01755 1.95 0.0517 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.211 on 580 degrees of freedom
## Multiple R-squared: 0.6021, Adjusted R-squared: 0.6007
## F-statistic: 438.8 on 2 and 580 DF, p-value: < 2.2e-16We ignore most of the output for now, and focus on the estimates of the coefficients. Namely, \(\beta_0 = 8.14199\), \(\beta_1 = 0.65130\) and \(\beta_2 = 0.03422\).
For comparison, these coefficients lead to a SSE of 25.8:
beta_0 <- 8.14199
beta_1 <- 0.65130
beta_2 <- 0.03422
log_houses %>%
mutate(
z_hat = beta_0 + beta_1 * log_sqft_living + beta_2 * log_sqft_lot,
resid = log_price - z_hat
) %>%
summarize(SSE = sum(resid^2))## SSE
## 1 25.8307This value is much smaller than the original value that we computed above!
The interested reader can check that if they change any of the coefficients by some small amount, then the SSE computed above will increase.
Let’s use plotly to visualize the regression plane.
We include the code for completeness, but do not explain the details of how to do this.
Plotly produces interactive plots that do not render well on paper, so we recommend running this code in R to see the results.
# Compute a grid of x,y values for the plane
x <- seq(min(log_houses$log_sqft_living),
max(log_houses$log_sqft_living),
length.out = 10
)
y <- seq(min(log_houses$log_sqft_lot),
max(log_houses$log_sqft_lot),
length.out = 10
)
grid <- expand.grid(x, y) %>%
rename(log_sqft_living = Var1, log_sqft_lot = Var2) %>%
select(starts_with("log")) %>%
modelr::add_predictions(price_mod)
z <- reshape2::acast(grid, log_sqft_lot ~ log_sqft_living,
value.var = "pred"
)plotly::plot_ly(
data = log_houses,
x = ~log_sqft_living,
y = ~log_sqft_lot,
z = ~log_price,
type = "scatter3d",
marker = list(size = 2, color = "black")
) %>%
plotly::add_trace(
x = ~x,
y = ~y,
z = ~z,
type = "surface"
)Let’s now examine the summary of lm when there are two explanatory variables. We will go through line by line, much as we did in the previous chapter, to explain what each value represents.
##
## Call:
## lm(formula = log_price ~ log_sqft_living + log_sqft_lot, data = log_houses)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.62790 -0.12085 -0.00189 0.12904 0.75567
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 8.14199 0.19922 40.87 <2e-16 ***
## log_sqft_living 0.65130 0.02303 28.29 <2e-16 ***
## log_sqft_lot 0.03422 0.01755 1.95 0.0517 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.211 on 580 degrees of freedom
## Multiple R-squared: 0.6021, Adjusted R-squared: 0.6007
## F-statistic: 438.8 on 2 and 580 DF, p-value: < 2.2e-16lm(formula = log_price ~ log_sqft_living + log_sqft_lot, data = log_houses)this gives the call tolmthat was used to create the model. It can be useful when you are comparing multiple models, or when a function creates a model for you without your typing it in directly.ResidualsThis gives the values of the estimated value of the response minus the actual value of the response. Many times, we will assume a model of \(y = \beta_0 + \beta_1 x + \beta_2 y + \epsilon\), where \(\epsilon\) is normal. In this case, we should expect that the residuals are symmetric about 0, so we would like to see that the median is about 0, the 1st and 3rd quartiles are about equal, and the min and max are about equal.CoefficientsThis gives the estimate for the coefficients, together with the standard error, \(t\)-value for use in a t-test of significance, and \(p\)-value. The \(p\)-value is for the test of \(H_0: \beta_i = 0\) versus \(H_a: \beta_i \not= 0\). In this instance, we reject the null hypotheses that the slope associated withlog_sqft_livingis zero at the \(\alpha = .05\) level, and fail to reject that the slope oflog_sqft_lotis zero at the \(\alpha = .05\) level. The \(\epsilon_i\) must be normal in order for these \(p\)-values to be accurate.Residual standard error. This can be computed from the residuals as follows:
## [1] 0.2110348Recall our rule of thumb that we divide by the sample size minus the number of estimated parameters when estimating \(\sigma\). In this case, there are three estimated parameters. The degrees of freedom is just the number of samples minus the number of parameters estimated.
Multiple R-squaredcan be interpreted as the percent of variance in the response that is explained by the model. It can be computed as follows:
var_response <- var(log_houses$log_price)
var_residuals <- var(residuals(price_mod))
(var_response - var_residuals) / var_response## [1] 0.6020751The Adjusted \(R^2\) uses the residual standard error (that has been adjusted for the number of parameters) rather than the raw variance of the residuals.
## [1] 0.600703The multiple \(R^2\) value will increase with the addition of more variables, where the adjusted \(R^2\) has a penalty for adding variables, so can either increase of decrease when a new variable is added.
F statisticthis is the test statistic used to test \(H_0: \beta_1 = \beta_2 = \cdots = 0\) versus not all of the slopes are 0. Note this does not include a test for the intercept. The \(\epsilon_i\) must be normal for this to be accurate. We compute the value via
ssresid <- sum(residuals(price_mod)^2)
ssresponse <- sum((log_houses$log_price - mean(log_houses$log_price))^2)
(ssresponse - ssresid) / 2 / (ssresid / (N - 3))## [1] 438.780813.2 Categorical variables
In this section we briefly explain hos R treats categorical variables in lm via an example. Let’s expand the ZIP codes in our model to include a zip code in the suburbs, which may have different characteristics. These are the same ZIP codes we used for data visualization in Section 7.3.1.
We again take the log of price, sqft_lot and sqft_living.
houses_2zips <- mutate(houses_2zips,
log_sqft_lot = log(sqft_lot),
log_sqft_living = log(sqft_living),
log_price = log(price)
)
houses_2zips$zipcode <- factor(houses_2zips$zipcode)
ggplot(houses_2zips, aes(
x = log_sqft_living, y = log_price,
color = zipcode
)) +
geom_point() +
geom_smooth(method = "lm")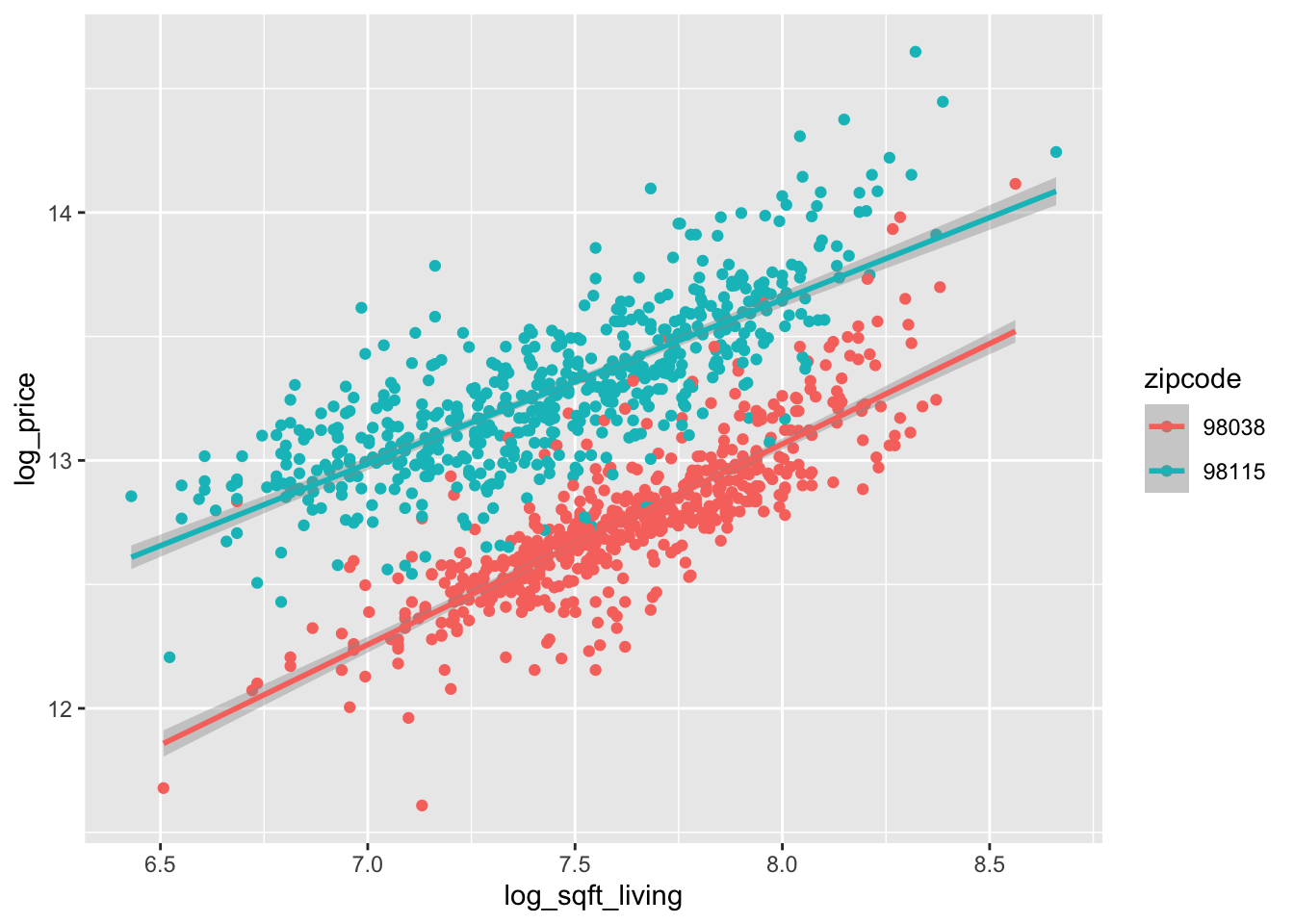
Based on this plot, it appears that 98038 and 98115 may have a similar slope of log_price when modeled by log_sqft_living, but the intercepts are very different. Let’s see how that pans out in lm.
It would be a mistake to treat zipcode as a numeric variable! We changed it to a factor above.
##
## Call:
## lm(formula = log_price ~ log_sqft_living + log_sqft_lot + zipcode,
## data = houses_2zips)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.96278 -0.09510 0.00084 0.09298 0.68666
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 6.82806 0.12150 56.20 <2e-16 ***
## log_sqft_living 0.67827 0.01562 43.43 <2e-16 ***
## log_sqft_lot 0.08329 0.00659 12.64 <2e-16 ***
## zipcode98115 0.69684 0.01182 58.97 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.1857 on 1169 degrees of freedom
## Multiple R-squared: 0.7999, Adjusted R-squared: 0.7994
## F-statistic: 1558 on 3 and 1169 DF, p-value: < 2.2e-16This says that all three variables are statistically significant. The way to interpret the zipcode coefficients is as follows. There is a “base” level of the factor, which can be obtained by typing levels(house$zipcode). The level that appears first is the “base” level. This means that if we have a house in ZIP code 98038 (the base level), we can compute its estimated log of price via 6.83 + 0.678 * sqft_living + 0.083 * sqft_lot. However, if we wish to compute the log price of a house in ZIP Code 98115, we would compute 6.83 + 0.678 * sqft_living + 0.083 * sqft_lot + 0.69684. That is, there is an additive adjustment that needs to be made when the house is in ZIP code 98115 rather than in 98038.
If you want to change the base level, you can do that as follows.
houses_2zips$zipcode <- factor(houses_2zips$zipcode,
levels = c("98115", "98038")
)
mod <- lm(log_price ~ log_sqft_living + log_sqft_lot + zipcode,
data = houses_2zips
)
summary(mod)##
## Call:
## lm(formula = log_price ~ log_sqft_living + log_sqft_lot + zipcode,
## data = houses_2zips)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.96278 -0.09510 0.00084 0.09298 0.68666
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 7.52489 0.11780 63.88 <2e-16 ***
## log_sqft_living 0.67827 0.01562 43.43 <2e-16 ***
## log_sqft_lot 0.08329 0.00659 12.64 <2e-16 ***
## zipcode98038 -0.69684 0.01182 -58.97 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.1857 on 1169 degrees of freedom
## Multiple R-squared: 0.7999, Adjusted R-squared: 0.7994
## F-statistic: 1558 on 3 and 1169 DF, p-value: < 2.2e-16Another possibility would be that we want to allow each ZIP code to have a different slope and intercept, instead of just different intercepts as above. To create this model, we add log_sqft_living:zipcode and log_sqft_lot:zipcode to the model. These indicate that we want to allow the slopes to depend on the level of the variable in zipcode.90
mod <- lm(log_price ~ log_sqft_living + log_sqft_lot + zipcode +
log_sqft_living:zipcode + log_sqft_lot:zipcode,
data = houses_2zips
)
summary(mod)##
## Call:
## lm(formula = log_price ~ log_sqft_living + log_sqft_lot + zipcode + ...
## Residuals:
## Min 1Q Median 3Q Max
## -0.94919 -0.09573 0.00371 0.09358 0.75567
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 8.14199 0.17375 46.861 < 2e-16 ***
## log_sqft_living 0.65130 0.02008 32.433 < 2e-16 ***
## log_sqft_lot 0.03422 0.01531 2.236 0.025558 *
## zipcode98038 -1.81575 0.25254 -7.190 1.16e-12 ***
## log_sqft_living:zipcode98038 0.08308 0.03175 2.617 0.008995 **
## log_sqft_lot:zipcode98038 0.05716 0.01695 3.373 0.000769 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.184 on 1167 degrees of freedom
## Multiple R-squared: 0.8038, Adjusted R-squared: 0.8029
## F-statistic: 956 on 5 and 1167 DF, p-value: < 2.2e-16Looking at the summary, we see that there is a small but statistically significant difference between the linear relationship of log of price to the other variables in the two zip codes. In order to interpret the estimates, we follow a similar path as above. To estimate the price of a house in ZIP code 98115 (recall we changed the base level above), we would compute 8.14199 + 0.65130 * log_sqft_living + 0.03422 * log_sqft_lot.
To estimate the expected value of the log of the price of a home in ZIP code 98038, we would compute 8.14199 - 1.81575 + (0.65130 + 0.08308) * log_sqft_living + (0.03422 + 0.05716) * log_sqft_lot. Of course, the predict function works on these models as well as on simple linear models, so we can check our results using predict as follows, for a house in ZIP code 98038 with log_sqft_living = 8 and log_sqft_lot = 9.
## 1
## 13.02381log_sqft_living <- 8
log_sqft_lot <- 9
8.14199 - 1.81575 + (0.65130 + 0.08308) * log_sqft_living +
(0.03422 + 0.05716) * log_sqft_lot # agrees with above## [1] 13.0237Finally, we note that predict has options for prediction intervals
and confidence intervals which work for multiple linear regression as well.
If we want a 95 percent prediction interval for the log price of a house with log_sqft_living = 8 and log_sqft_lot = 9,
we see that it is \([12.66, 13.39]\), which corresponds roughly to \([315000 ,651000]\) in the non-log scale.
exp(predict(mod,
newdata = data.frame(
log_sqft_living = 8,
log_sqft_lot = 9, zipcode = "98038"
),
interval = "pre"
))## fit lwr upr
## 1 453072.1 315496.9 650638.213.3 Variable selection
One of the goals of linear regression is to be able to choose a parsimonious set of explanatory variables.
There are many ways of achieving that, and we present one using \(p\)-values in this section. We consider the data set fosdata::conversation , adapted from a psychological study by Manson et. al.91
This study had 210 university students participate in a 10 minute conversation in small groups. The participants also provided various demographic information, and took a test to indicate whether they suffered from subclinical psychopathy. The goal was to determine whether and/or how conversational items such as the proportion of words spoken depend on the demographic data. You can read how the authors of the study analyzed the data in the paper linked above. For us, we want to use this data set to illustrate variable selection, so we are going to model the proportion of words spoken on the variables in the data set. The variables split naturally into two groups: demographic data and conversational data. The conversational data measures the number of interruptions, the number of times that a person started a new topic on a per word basis, and the proportion of times a person started a new topic. The demographic data includes data on shirt type, gender, median income of major, prestige of major, fighting ability (as estimated based on a picture), etc.
A few things to notice about this data set. First, the character data should be encoded as factors, as should gender.
converse$oldest <- factor(converse$oldest)
converse$gender <- factor(converse$gender)
summary(converse)We do not print out the entire summary, but you can see that many of the variables have been standardized, so as to have approximately mean 0 and standard deviation 1, and that oldest has 72 missing values. We need to decide what to do with that variable, but since it isn’t well documented in the paper, let’s just remove it. We also remove all of the information related to the prisoner’s dilemma part of the experiment. That part of the experiment by necessity happened after the conversations, so it seems to make more sense to model the prisoner’s dilemma outcomes on the conversation than the other way around.
Let’s examine some plots. Here, we pick a few of the explanatory variables to plot versus the response.
ggplot(converse, aes(x = f1_psychopathy, y = proportion_words)) +
geom_point() +
geom_smooth(method = "lm")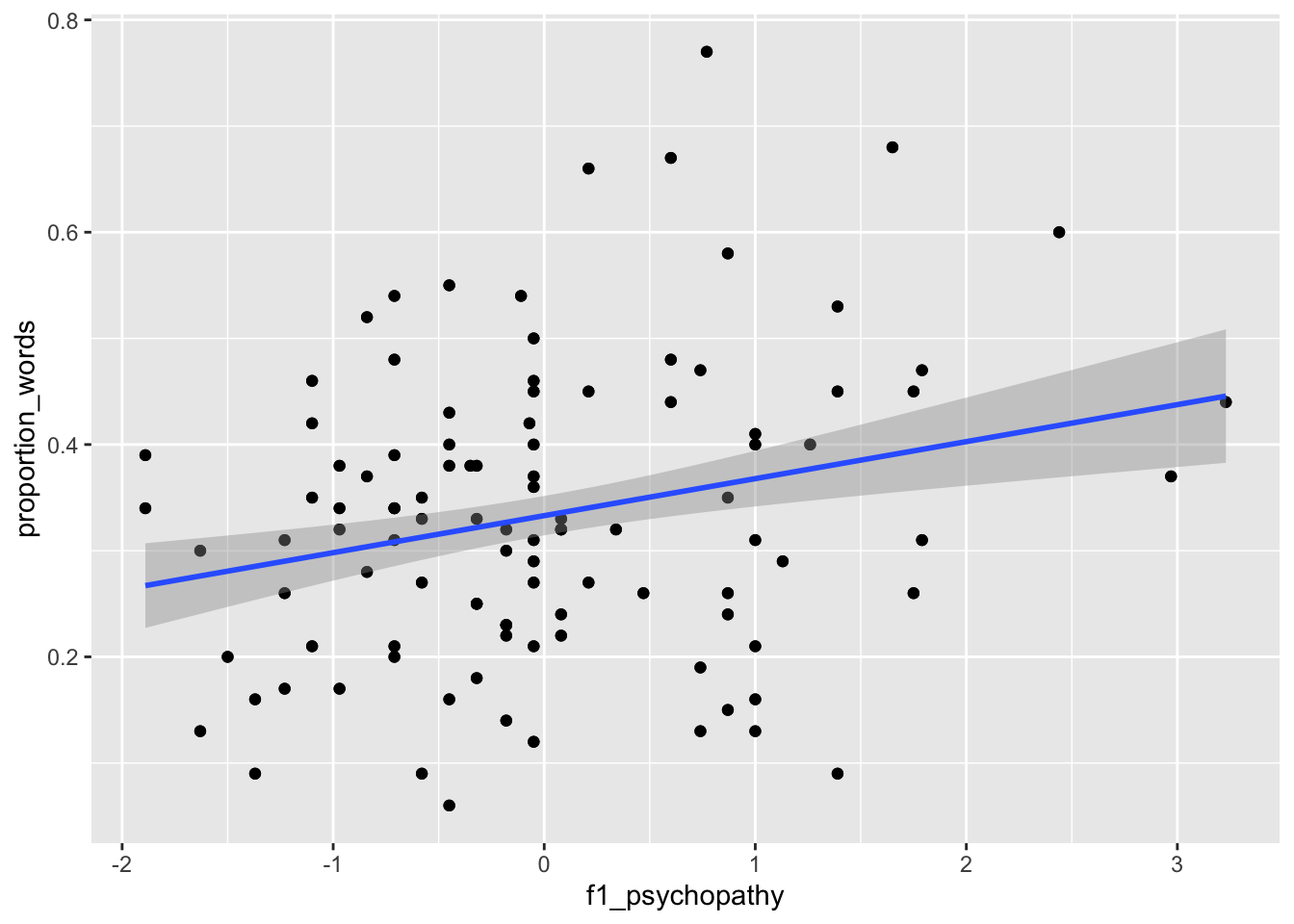
ggplot(converse, aes(x = interruptions_per_min, y = proportion_words)) +
geom_point() +
geom_smooth(method = "lm")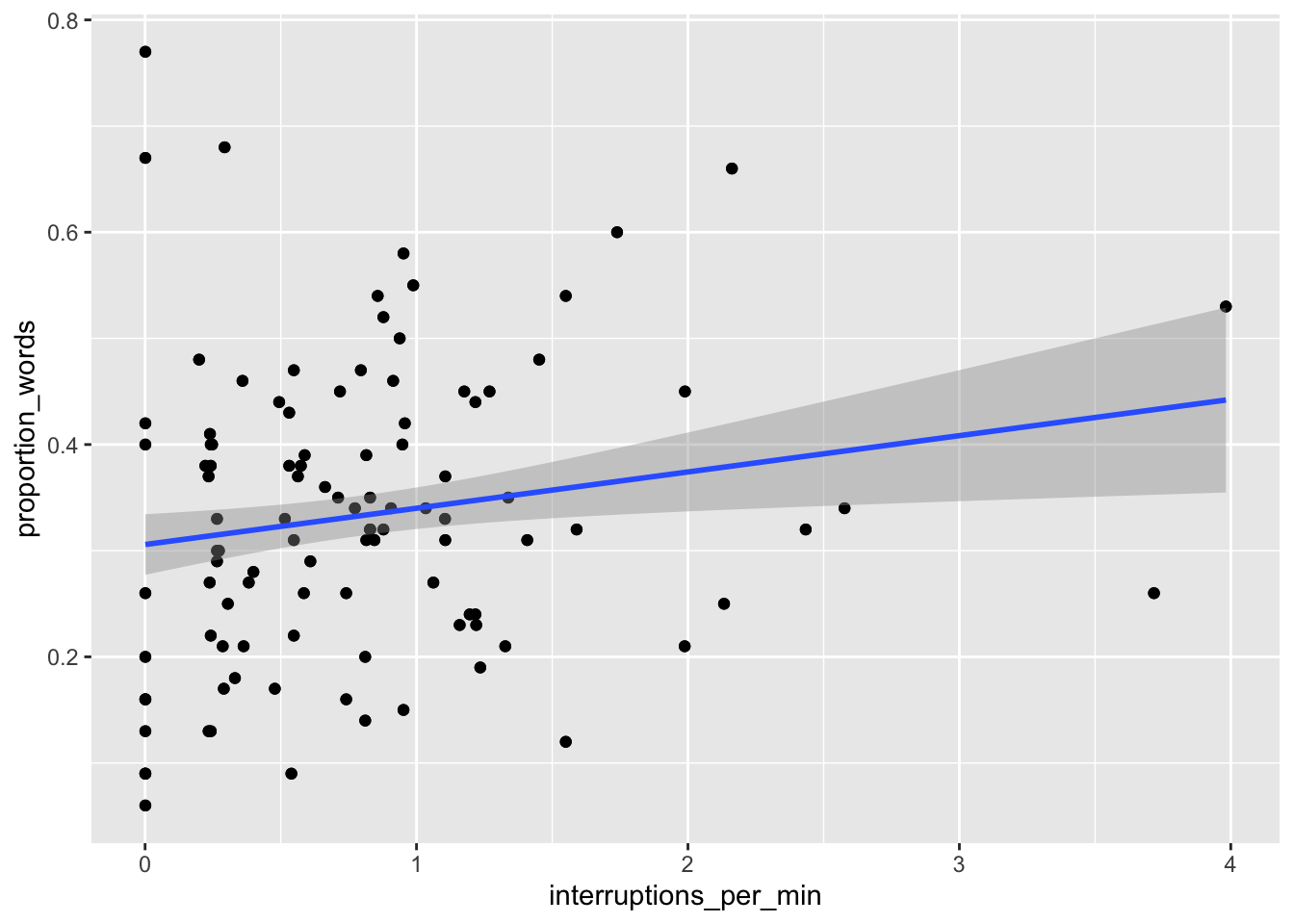
We can see that neither of the two above variables have strong predictive ability for the response by themselves, but perhaps with all of the variables, things will look better.
##
## Call:
## lm(formula = proportion_words ~ ., data = converse)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.183163 -0.040952 0.002491 0.044088 0.188120
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.2959771 0.0305151 9.699 < 2e-16 ***
## gender1 0.0051863 0.0118649 0.437 0.66252
## f1_psychopathy -0.1579680 0.4577533 -0.345 0.73040
## f2_psychopathy -0.0980854 0.2457922 -0.399 0.69029
## total_psychopathy 0.2237067 0.5963622 0.375 0.70799
## attractiveness -0.0022099 0.0055057 -0.401 0.68859
## fighting_ability -0.0286368 0.0086793 -3.299 0.00115 **
## strength 0.0186639 0.0085663 2.179 0.03057 *
## height -0.0057262 0.0055482 -1.032 0.30333
## median_income 0.0001271 0.0002090 0.608 0.54379
## highest_class_rank 0.0242887 0.0115059 2.111 0.03607 *
## major_presige -0.0005630 0.0005683 -0.991 0.32313
## dyad_status_difference -0.0178477 0.0070620 -2.527 0.01230 *
## proportion_seque...rts 0.4855446 0.0409178 11.866 < 2e-16 ***
## interruptions_per_min 0.1076160 0.0144719 7.436 3.35e-12 ***
## sequence_starts_...100 -0.0493571 0.0061830 -7.983 1.28e-13 ***
## interruptions_pe...100 -0.0708835 0.0094431 -7.506 2.22e-12 ***
## affect_words -0.0039201 0.0027227 -1.440 0.15155
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.07303 on 192 degrees of freedom
## Multiple R-squared: 0.751, Adjusted R-squared: 0.729
## F-statistic: 34.07 on 17 and 192 DF, p-value: < 2.2e-16A couple of things to notice. First, two of the other three conversation variables are significant when predicting the proportion of words spoken. Second, the multiple \(R^2\) is .751, which means that about 75 percent of the variance in the proportion of words spoken is explained by the explanatory variables. Let’s start removing variables in groups. First, we have the three psychopathy variables. We start by removing them one at a time. We will use the update command, which takes as its first argument the model that we are updating, and the argument is the update that we are making. The form of the second argument we will be using is ~ . - variable_name. For reasons of space, we do not show the entire output at each step. You won’t need to do this when doing your own analysis.
mod2 <- update(mod, ~ . - f1_psychopathy)
mod2 %>%
broom::tidy() %>%
mutate(across(where(is.numeric), ~ round(., 3))) %>%
filter(stringr::str_detect(term, "psycho|Inter")) %>%
knitr::kable()| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 0.295 | 0.030 | 9.745 | 0.000 |
| f2_psychopathy | -0.013 | 0.008 | -1.656 | 0.099 |
| total_psychopathy | 0.018 | 0.009 | 2.102 | 0.037 |
## [1] 0.7508613## [1] 0.7302073We see that after removing f1_psychopathy, the variabletotal_psychopathy has coefficient significantly different from 0 (\(p = .037\)), while f2_psychopathy is not significant at the \(\alpha = .05\) level. Also, note that the \(R^2\) did not decrease much when we removed f1_psychopathy, and the adjusted \(R^2\) increased from .729 to .730.. These are good signs that it is OK to remove f1_psychopathy. It is common to see large changes in significance when removing variables that are linked or correlated in some way. We continue removing the variables linked to psychopathy. So, we leave those two variables in for now, and continue removing variables. It seems that gender can be removed.
mod3 <- update(mod2, ~ . - f2_psychopathy)
mod3 %>%
broom::tidy() %>%
mutate(across(where(is.numeric), ~ round(., 3))) %>%
filter(stringr::str_detect(term, "psycho|Inter")) %>%
knitr::kable()| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 0.296 | 0.030 | 9.731 | 0.000 |
| total_psychopathy | 0.007 | 0.006 | 1.293 | 0.198 |
## [1] 0.7473232## [1] 0.7277863Here we get conflicting information about whether to remove f2_psychopathy. The variable was not significant at the \(\alpha = .05\) level, but the adjusted \(R^2\) decreases from .730 to .728 when we remove the variable. Different variable selection techniques would do different things at this point. However, we are using \(p\)-values right now, so we continue by removing the last of the psychopathy variables (since it is not significant).
##
## Call:
## lm(formula = proportion_words ~ gender + attractiveness + fighting_a...
## Residuals:
## Min 1Q Median 3Q Max
## -0.180736 -0.038420 -0.000994 0.043020 0.202987
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.2933090 0.0303800 9.655 < 2e-16 ***
## gender1 -0.0037965 0.0108581 -0.350 0.726983
## attractiveness -0.0028329 0.0055096 -0.514 0.607706
## fighting_ability -0.0304228 0.0083761 -3.632 0.000359 ***
## strength 0.0211761 0.0083132 2.547 0.011628 *
## height -0.0082203 0.0053578 -1.534 0.126585
## median_income 0.0001295 0.0002088 0.620 0.535722
## highest_class_rank 0.0244283 0.0114397 2.135 0.033976 *
## major_presige -0.0004255 0.0005441 -0.782 0.435162
## dyad_status_difference -0.0153037 0.0069836 -2.191 0.029610 *
## proportion_seque...rts 0.5013595 0.0399347 12.554 < 2e-16 ***
## interruptions_per_min 0.1138530 0.0141768 8.031 8.97e-14 ***
## sequence_starts_...100 -0.0481660 0.0061777 -7.797 3.73e-13 ***
## interruptions_pe...100 -0.0731355 0.0093695 -7.806 3.53e-13 ***
## affect_words -0.0050216 0.0026643 -1.885 0.060946 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.07331 on 195 degrees of freedom
## Multiple R-squared: 0.7451, Adjusted R-squared: 0.7268
## F-statistic: 40.72 on 14 and 195 DF, p-value: < 2.2e-16The adjusted \(R^2\) fell again, this time to .727. Now that we have removed all of the variables related to psychopathy, we can check to see whether as a group they are significant. That is, we will test \(H_0\) the coefficients of the psychopathy variables are all 0 versus \(H_a\) not all of the coefficients are zero.
## Analysis of Variance Table
##
## Model 1: proportion_words ~ gender + attractiveness + fighting_ability...
## Model 2: proportion_words ~ gender + f1_psychopathy + f2_psychopathy +...
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 195 1.0481
## 2 192 1.0240 3 0.024138 1.5087 0.2136We get a \(p\)-value of .2136, so we do not reject the null hypothesis that all of the coefficients are zero. If we had rejected the null hypothesis, then we would have needed to add back in one or more of the variables to the model. It is important when doing stepwise variable selection that you periodically check to see whether you need to add back in one or more of the variables that you removed. Next, we remove the variables that describe the person, such as attractiveness or strength. Again, we go in order of \(p\)-values within that group of variables.
mod5 <- update(mod4, ~ . - gender)
mod6 <- update(mod5, ~ . - attractiveness)
mod7 <- update(mod6, ~ . - median_income)
summary(mod7)##
## Call:
## lm(formula = proportion_words ~ fighting_ability + strength + ...
## Residuals:
## Min 1Q Median 3Q Max
## -0.181215 -0.037676 -0.000914 0.042230 0.202280
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.3024526 0.0214860 14.077 < 2e-16 ***
## fighting_ability -0.0300948 0.0082729 -3.638 0.000351 ***
## strength 0.0207458 0.0082203 2.524 0.012397 *
## height -0.0080111 0.0052692 -1.520 0.130012
## highest_class_rank 0.0243218 0.0112871 2.155 0.032382 *
## major_presige -0.0004917 0.0005239 -0.939 0.349084
## dyad_status_difference -0.0144526 0.0067093 -2.154 0.032439 *
## proportion_seque...rts 0.5008202 0.0393863 12.716 < 2e-16 ***
## interruptions_per_min 0.1127051 0.0138752 8.123 4.81e-14 ***
## sequence_starts_...100 -0.0488678 0.0060006 -8.144 4.22e-14 ***
## interruptions_pe...100 -0.0725000 0.0092657 -7.825 2.99e-13 ***
## affect_words -0.0048069 0.0025190 -1.908 0.057807 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.07289 on 198 degrees of freedom
## Multiple R-squared: 0.7442, Adjusted R-squared: 0.73
## F-statistic: 52.37 on 11 and 198 DF, p-value: < 2.2e-16## Analysis of Variance Table
##
## Model 1: proportion_words ~ fighting_ability + strength + height + hig...
## Model 2: proportion_words ~ gender + attractiveness + fighting_ability...
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 198 1.0520
## 2 195 1.0481 3 0.0038477 0.2386 0.8693After removing those three variables, the adjusted \(R^2\) is back up to 0.73, and ANOVA indicates it is not necessary to put any of the variables back in. We continue.
mod8 <- update(mod7, ~ . - major_presige)
mod9 <- update(mod8, ~ . - height)
mod10 <- update(mod9, ~ . - dyad_status_difference)
anova(mod10, mod7)## Analysis of Variance Table
##
## Model 1: proportion_words ~ fighting_ability + strength + highest_clas...
## Model 2: proportion_words ~ fighting_ability + strength + height + hig...
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 201 1.0893
## 2 198 1.0520 3 0.037357 2.3438 0.07427 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1##
## Call:
## lm(formula = proportion_words ~ fighting_ability + strength + ...
## Residuals:
## Min 1Q Median 3Q Max
## -0.192950 -0.041033 0.003062 0.042783 0.211376
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.295980 0.020301 14.580 < 2e-16 ***
## fighting_ability -0.033053 0.008021 -4.121 5.52e-05 ***
## strength 0.021673 0.008193 2.645 0.00881 **
## highest_class_rank 0.023024 0.011185 2.058 0.04084 *
## proportion_seque...rts 0.502863 0.039371 12.772 < 2e-16 ***
## interruptions_per_min 0.113354 0.013994 8.100 5.22e-14 ***
## sequence_starts_...100 -0.048002 0.005906 -8.128 4.41e-14 ***
## interruptions_pe...100 -0.072946 0.009305 -7.840 2.59e-13 ***
## affect_words -0.005876 0.002418 -2.430 0.01597 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.07362 on 201 degrees of freedom
## Multiple R-squared: 0.7351, Adjusted R-squared: 0.7246
## F-statistic: 69.73 on 8 and 201 DF, p-value: < 2.2e-16The adjusted \(R^2\) has fallen back down to .7246, but that is not a big price to pay for removing so many variables from the model. ANOVA does not reject \(H_0\) that all the coefficients of the variables that we removed are zero at the \(\alpha = .05\) level. We are left with 8 significant explanatory variables.
Let’s check the diagnostic plots.
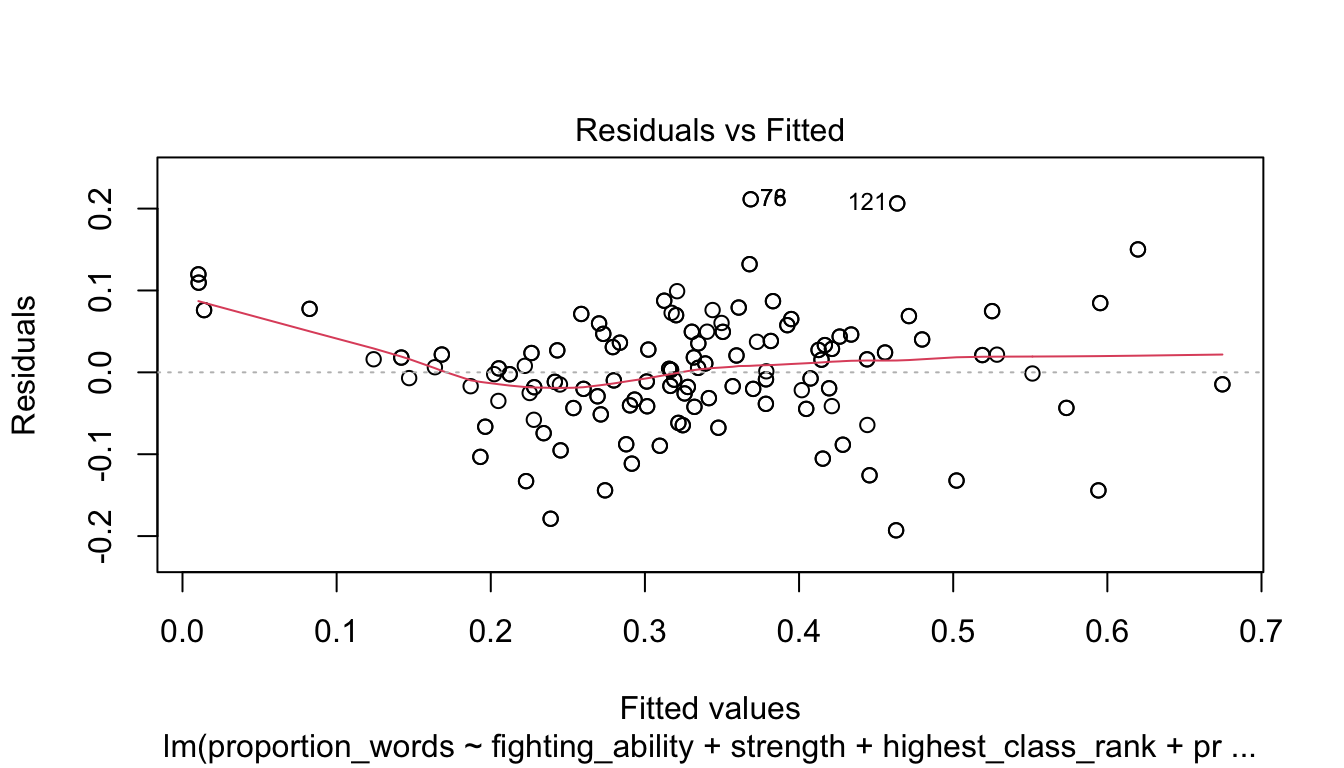
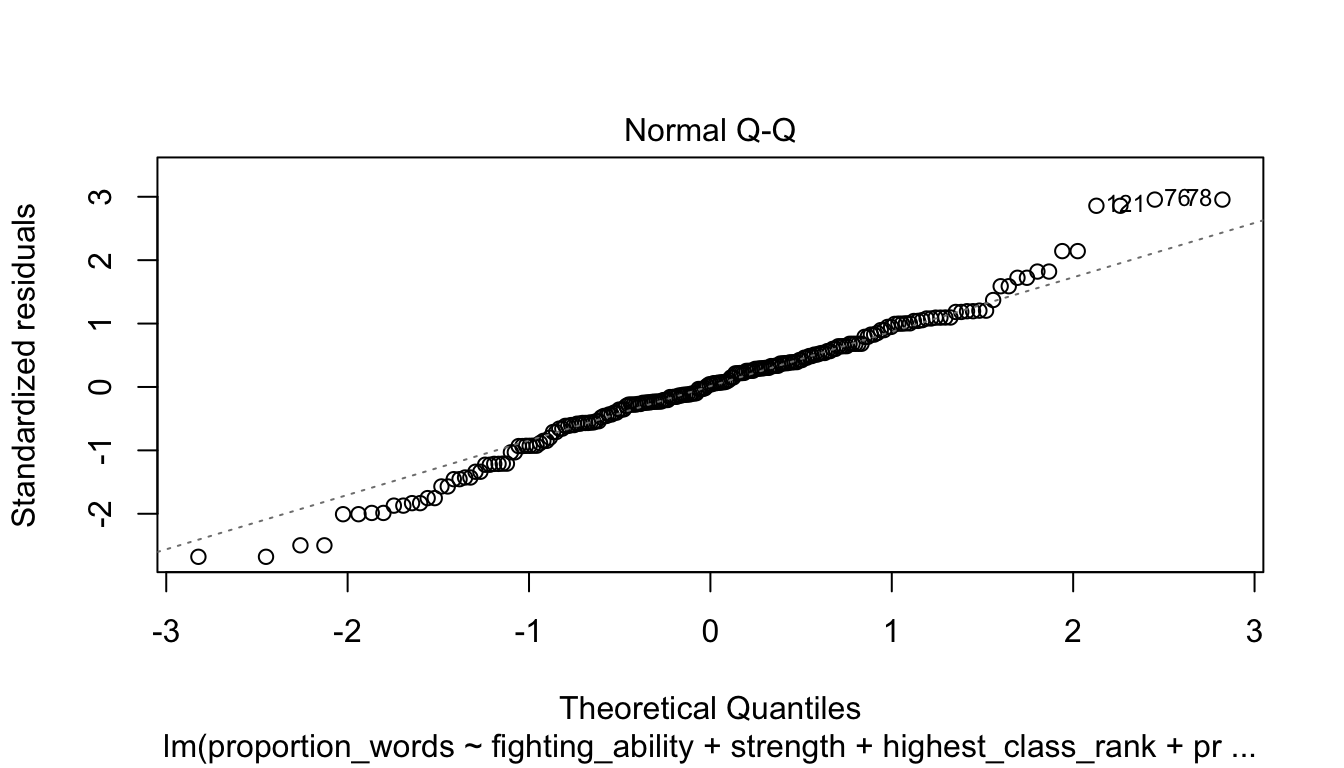
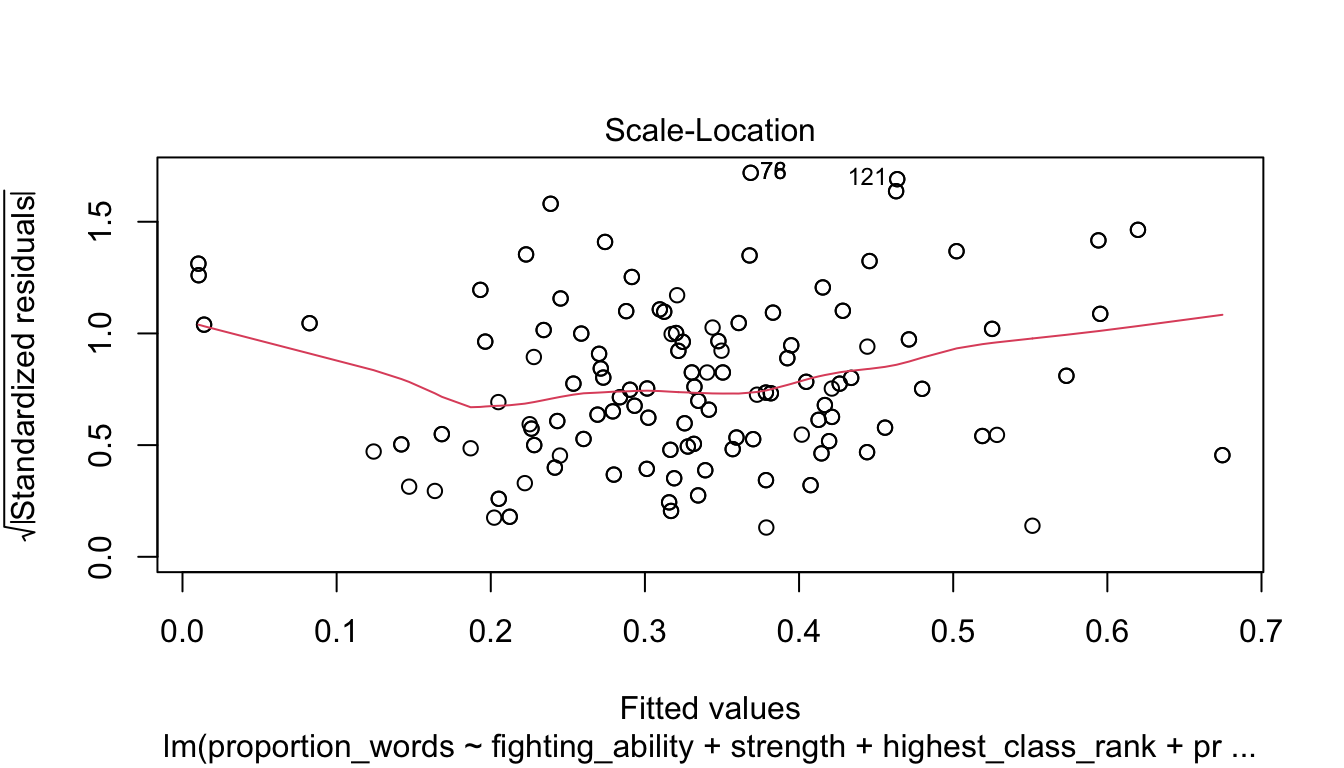
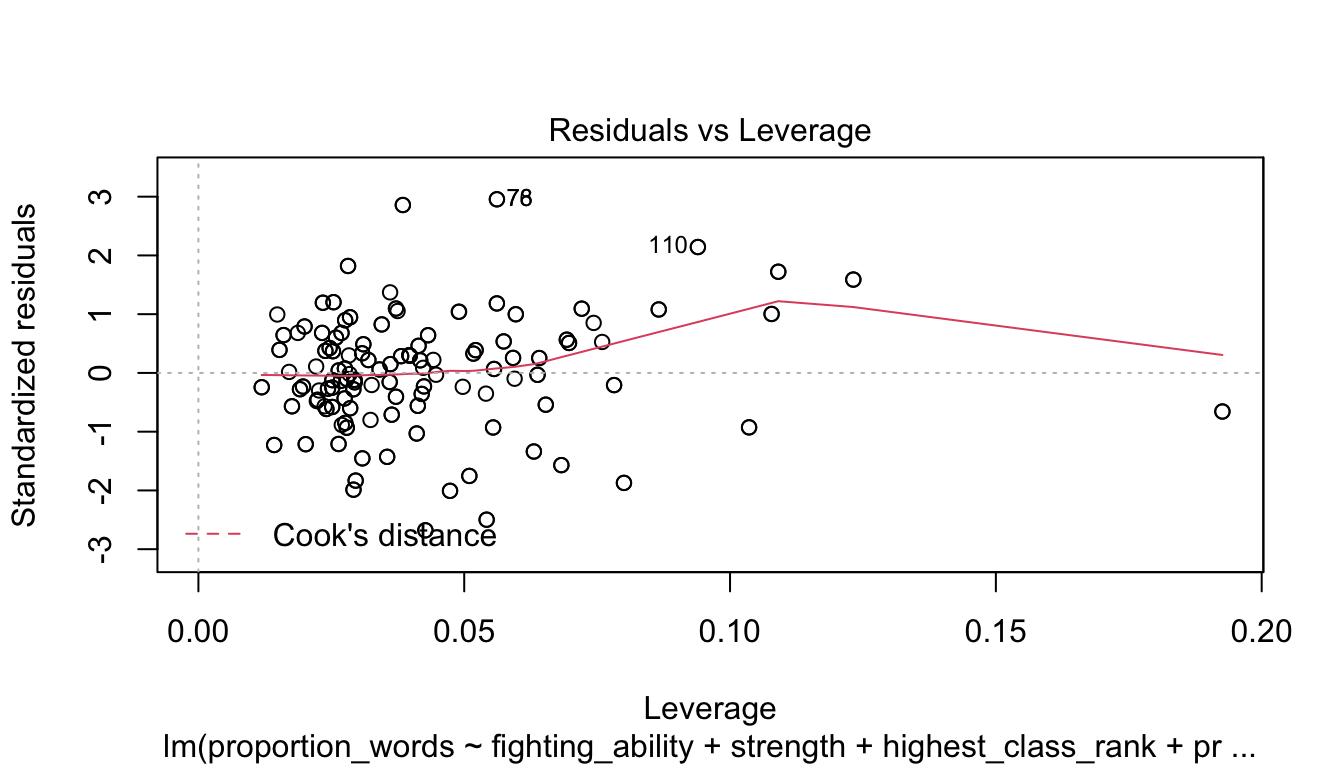
These look pretty good, except that there may be 3-4 values that don’t seem to be following the rest of the trend. See for example the residuals versus fitted plot with the 4 values on the left.
To summarize, here is a method for doing stepwise regression using \(p\)-values.
- Group the variables into variables that naturally belong together, if possible.
- Remove the non-significant variables in a group, checking each time to see whether the significance of other variables has changed. If a variable is categorical, then keep the variable if any of the levels of the variable are significant.
- Use
anovato see whether any of the variables that were removed are significant. If so, test one-by-one to try to find one to put back in the model. - Move to the next group of variables and continue until all of the variables are significant.
Doing stepwise regression like this is not without its detractors. One issue is that the final model is very much dependent on the order in which we proceed. If we had removed f2_psychopathy first, how would that have affected things? Can we justify our decision to remove f1_psychopathy first? Certainly not. The resulting model that we obtained is simply a model for the response. We are not claiming that it is the best one. It is also very possible that the variables that we removed are also good predictors of the response. We should not make the conclusion that the other variables aren’t related.
That being said, stepwise regression for variable selection is quite common in practice, which is why we include it in this book.
The technique we used above was based on \(p\)-values. One can also use other measures of goodness of fit of a model to perform stepwise regression. A common one is the Akaike Information Criterion, which we do not explain here, but we show how to use it to find a parsimonious model. The implementation of this is nice because at each decision point, the algorithm tests what would happen if we add back in any of the variables that we have removed as well as what would happen if we remove any of the variables that we currently have.
##
## Call:
## lm(formula = proportion_words ~ f2_psychopathy + total_psychopathy +...
## Residuals:
## Min 1Q Median 3Q Max
## -0.189974 -0.036895 0.000572 0.044182 0.190963
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.304026 0.020184 15.063 < 2e-16 ***
## f2_psychopathy -0.013849 0.007641 -1.812 0.071434 .
## total_psychopathy 0.018827 0.007876 2.390 0.017773 *
## fighting_ability -0.032058 0.008090 -3.963 0.000103 ***
## strength 0.020196 0.008139 2.481 0.013919 *
## highest_class_rank 0.024566 0.011126 2.208 0.028394 *
## dyad_status_difference -0.015754 0.006712 -2.347 0.019909 *
## proportion_seque...rts 0.479287 0.039657 12.086 < 2e-16 ***
## interruptions_per_min 0.106362 0.014115 7.536 1.70e-12 ***
## sequence_starts_...100 -0.047826 0.005855 -8.168 3.63e-14 ***
## interruptions_pe...100 -0.071390 0.009274 -7.698 6.42e-13 ***
## affect_words -0.005436 0.002393 -2.272 0.024170 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.07246 on 198 degrees of freedom
## Multiple R-squared: 0.7472, Adjusted R-squared: 0.7332
## F-statistic: 53.21 on 11 and 198 DF, p-value: < 2.2e-16In this case, the AIC derived model includes all of the variables that we included, plus f2_psychopathy, total_psychopathy, and dyad_status_difference. These variables do not all pass the \(p\)-value test, but notice that the adjusted \(R^2\) of the AIC derived model is higher than the adjusted \(R^2\) of the model that we chose. The two models have their pros and cons, and as is often the case, there is not just one final model that could make sense.
Vignette: External data formats
By this point in the book, we hope that you are comfortable doing many tasks in R. We like to imagine that when someone gives you data in the future, your first thought will be to open it up in R. Unfortunately, not everyone is as civilized as we are, and they may have saved their data in a format that doesn’t interact well with R. This vignette gives some guidance for what to do in that case.
As a general recommendation, never make modifications directly to the original data. That data should stay in the exact same state, and ideally, you would document every change and decision that you make while getting the data ready to be read into R. In particular, never write over the original data set.
Text formats
Text data is data stored as a sequence of simple characters (ASCII or Unicode, usually). It is easy to inspect and manipulate with any text editor. On the other hand, it typically does not include formatting or metadata (information about the data), and may be larger in size than some compressed formats. Despite, or really because of their simplicity, textual data formats have proven to be the best storage format for interoperability and resistance to loss over time. If you have important data to release to the world, do the world a favor make it available in a text format.
The most popular textual format is “comma separated values”, or CSV.
CSV files have the .csv extension, and can be read with read.csv(file = "your_file_name.csv").
Within a CSV file, each record is a row, and within that row data is separated into fields.
The first row of a CSV file should contain the names of the fields, or variables.
If it doesn’t, specify that header = FALSE and R will assign generic variable names to your columns.
The default field separator is a comma, hence the C in CSV. You may specify a different separator to read.csv.
For example, files with the .tsv extension are tab separated and need the sep = "\t" option
Other text formats with records in rows and variables in columns can be read with read.table, and in fact
read.csv is just a wrapper that eventually calls read.table. Another possible approach for reading
text files is with scan, which reads typed data. The readr package, part of the tidyverse, is also powerful tool.
JSON is a text based format that is growing in popularity. Unlike row and column formatted table data, JSON is
heirarchical, allowing for complicated object structures and relationships. Since R works most naturally with
rectangular data, dealing with JSON is inherently challenging. R does provide packages (rjson, jsonlite) that will
read JSON formats, but some data manipulation will almost certainly be required after loading.
Spreadsheets
Excel spreadsheet data is stored in files with .xls or .xlsx extensions.
The readxl package provides
readxl::read_xls("your_file_name.xls", sheet = 1) or readxl::read_xlsx(your_file_name.xlsx, sheet = 1) to import Excel data.
Unfortunately, the readxl commands will only work well if the file was created by someone who understands the importance of rectangular data.
Many, if not most, of the data that is stored in Excel format has data and metadata stored in the file itself.
For example, cells might be merged for formatting, there may be 5 rows of headers describing the variables,
or there might be cells performing computations - those aren’t data!
Working with complex Excel spreadsheets inside of R is possible, but quite challenging.
It is almost always easier to open the file in a spreadsheet program such as Google Sheets, Excel, or LibreOffice,
and perform some pre-processing by hand.
After cleaning the data in the spreadsheet, save it to a .csv file which can be read into R.
Unfortunately, the by hand nature of this process is not reproducible.
At a minimum, keep a copy of the original Excel file.
Binary formats
Statistical packages all have their own specialized file formats. Unlike text data,
without a system designed to interpret the format you have little hope of getting at the data.
Fortunately, R has packages that can read most specialized formats. The haven package is part of the
tidyverse, and provides commands read_dat(), read_stata(), read_spss() and read_sas() among others.
These read .dat (MATLAB), .dta (Stata), .sav (SPSS), and .sas7bdat (SAS) files.
The haven package usually works well, though the format of the imported data can seem a bit odd.
If the data file extension is .RData or .Rda, your file is likely an R Data file and can be loaded into R via load("file_name.RData").
This is the easiest case to deal with for an R user!
For less common formats, either convert to a format that R can read, or look for a speciality package that can read your data type natively. Chances are, there is one out there.
Exercises
Exercises 13.1 - 13.4 require material through Section 13.1.
Consider the adipose data set in the fosdata package.
The goal is to estimate the visceral adipose tissue amount in patients, based on the other measurements.
The authors of the study did separate analyses for males and females; in this exercise, we consider only male subjects.
- Model
vatonwaist_cmandstature_cmand examine the residuals. - Model
log(vat)onwaist_cmandstature_cmand examine the residuals. - Which model would you choose?
Continuing from Exercise 13.1, in this exercise we consider only female patients.
- The
vatmeasurement of some females was compromised. Remove any female whosevatvalue is less than or equal to 5. - Model
log(vat)onwaist_cmandstature_cm, solog(vat) = beta0 + beta1 * waist_cm + beta2 * stature_cm. - What is the \(p\)-value for the test of \(H_0: \beta_1 = \beta_2 = 0\) versus the alternative that at least one is not zero?
- What is the \(p\)-value for the test \(H_0: \beta_1 = 0\) versus \(H_a: \beta_1 \not= 0\)?
- What is a 95 percent confidence interval for
beta_2? - Find a 95 percent prediction interval for
vatfor a new patient who presents withwaist_cm = 70andstature_cm = 170.
In this problem, we will use simulation to show that the \(F\) statistic computed by lm follows the \(F\) distribution.
- Create a data frame where
x1andx2are uniformly distributed on \([-2, 2]\), and \(y = 2 + \epsilon\), where \(\epsilon\) is a standard normal random variable. - Use
lmto create a linear model of \(y = \beta_0 + \beta_1 x_1 + \beta_2 x_2\). - Use
summary(mod)$fstatistic[1]to pull out the \(F\) statistic. - Put inside
replicate, and create 2000 samples of the \(F\) statistic (for different response variables). - Create a histogram of the \(F\) statistic and compare to the pdf of an \(F\) random variable with the correct degrees of freedom. (There should be 2 numerator degrees of freedom and \(N\) - 3 denominator degrees of freedom.)
In this problem, we examine robustness to normality in lm.
- Create a data frame of 20 observations where
x1andx2are uniformly distributed on \([-2, 2]\), and \(y = 1 + 2 * x_1 + \epsilon\), where \(\epsilon\) is exponential with rate 1. - Use
lmto create a linear model of \(y = \beta_0 + \beta_1 x_1 + \beta_2 x_2\), and usesummary(mod)$coefficients[3,4]to pull out the \(p\)-value associated with a test of \(H_0: \beta_2 = 0\) versus \(H_a: \beta_2 \not= 0\). - Replicate the above code and estimate the proportion of times that the \(p\)-value is less than .05.
- How far off is the value from what you would expect to get from the theory?
Exercise 13.5 requires material through Section 13.2.
Consider the data set fosdata::fish, . This data set consists of the weight, species, and 5 other measurements of fish.
- The
speciesvariable is coded as integer. Is this the appropriate coding for doing regression? - Find a linear model of
weightonspecies,length1and the interaction betweenspeciesandlength1. - What is the expected weight of a fish that is species 3 and length1 24.1?
- What is the expected difference of weight between a fish of species 3 and length1 of 22.1 and a fish of species 3 and length1 of 23.1?
Exercises 13.6 - 13.12 require material through Section 13.3.
Consider again the conversation data set in the fosdata package.
Suppose that we wanted to model the proportion of words spoken on variables that would be available before the conversation.
- Find a parsimonious model of the proportion of words spoken on
gender,f1_psychopathy,f2_psychopathy,total_psychopathy,attractiveness,fighting_ability,strength,height,median_income,highest_class_rank,major_presigeanddyad_status_difference. - What is the \(R^2\) value? Discuss.
Consider the adipose data set in the fosdata package.
- Create a parsimonious model of
vaton the other numeric variables for male patients. Which variables are kept? - What is the expected value of
vatfor a male patient who presents withage = 20,ldl = 2,hdl = 1.5,trig = 0.6,glucose = 4.6,stature_cm = 170,waist_cm = 72,hips_cm = 76andbmi = 19?
Consider the data set fosdata::cigs_small.
The Federal Trade Commission tested 1200 brands of cigarettes in 2000, and reported the carbon monoxide, nicotine, and tar content along with other identifying variables.
The data in small_cigs contains one randomly selected cigarette from each brand that made a 100 cigarette in 2000. Find a parsimonious model of co on the variables nic, tar, pack and menthol.
Consider the data set fosdata::fish, . This data set consists of the weight, species, and 5 other measurements of fish. Find a parsimonious model of weight on the other variables. Note that there are many missing values for gender.
Consider the data set tlc in the ISwR package.
Model the variable tlc inside the data set tlc on the other variables. Include plots and examine the residuals.
Consider the cystfibr data set in the ISwR package.
Find a parsimonious linear model of the variable pemax on the other variables. Be sure to read about the variables so that you can guess which variables might be grouped together.
Consider the seoulweather data set in the fosdata package. We wish to model the error in the next day forecast max temperature on the variables that we knew on the present day. (That is, all variables except for next_tmax and next_tmin.)
- Find a parsimonious model of the error on the other variables.
- What percentage of the error in the next day prediction is explained by your model?
Throughout this chapter, we truncate the output of
summary(mod)in two ways. If the call is long, we only show the first line of the call. If the variable names are long enough that would cause the Coefficientes table to wrap, we truncate the variable names.↩︎Manson JH, Gervais MM, Fessler DMT, Kline MA (2014) Subclinical Primary Psychopathy, but Not Physical Formidability or Attractiveness, Predicts Conversational Dominance in a Zero-Acquaintance Situation. PLoS ONE 9(11): e113135. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0113135↩︎